
The enzyme will use ADP5/6 of the time, and CDP 1/6 of the time. For example, consider a 5:1 mixtureof A:C. If two NDPs are mixed in a knownratio, polynucleotide phosphorylase will make a mixed co‑polymer in whichnucleotide is incorporated at a frequency proportional to its presence in theoriginal mixture.ī. Poly(G)programmed synthesis of poly‑Gly GGG encodes Gly.Ī. Poly(C)programmed synthesis of poly‑Pro CCC encodes Pro. Likewise, poly(A) programmed synthesisof poly‑Lys AAA encodesLys. coli lysates), results in a newly synthesizedpolypeptide which is a polymer of polyphenylalanine.ĭ. Addition of poly(U) to an in vitrotranslation system (e.g. If you provide only UDP as asubstrate for polynucleotide phosphorylase, the product will be a homopolymerpoly(U).ī. Homopolymers program synthesis ofspecfic homo‑polypeptidesĪ. However, in a cell-free system, the forward reaction isvery useful for making random RNA polymers.ģ. The physiological function ofpolynucleotide phosphorylase is to catalyze the reverse reaction, which is usedin RNA degradation.
Ochoa isolatedthe enzyme polynucleotide phosphorylase, and showed that it was capableof linking nucleoside di phosphates(NDPs) into polymers of NMPs (RNA) in a reversible reaction. The ability to synthesize randompolynucleotides was another key development to allow the experiments todecipher the code. Mammalian (rabbit) reticulocytes: ribosomes actively making lots ofglobin.Ģ. This ability to carry out translationin vitro was one of the technical advances needed to allow investigators todetermine the genetic code.Ī. Several different cell‑freesystems have been developed that catalyze protein synthesis.
The wild-type reading frame is restored after the 3 rddeletion (or insertion).ġ. Combinations of three different single nucleotide deletions(or insertions), each of which has a loss-of-function phenotype individually,can restore substantial function to a gene. In the latter case, the reading frame is maintained, with aninsertion or deletion of an amino acid at one site. But those that addor delete three nucleotides have little or no effect. Length‑altering mutations thatadd or delete one or two nucleotides have severe defective phenotype (theychange the reading frame, so the entire amino acid sequence after the mutationis altered.).
#Large hydrophobic amino acids code
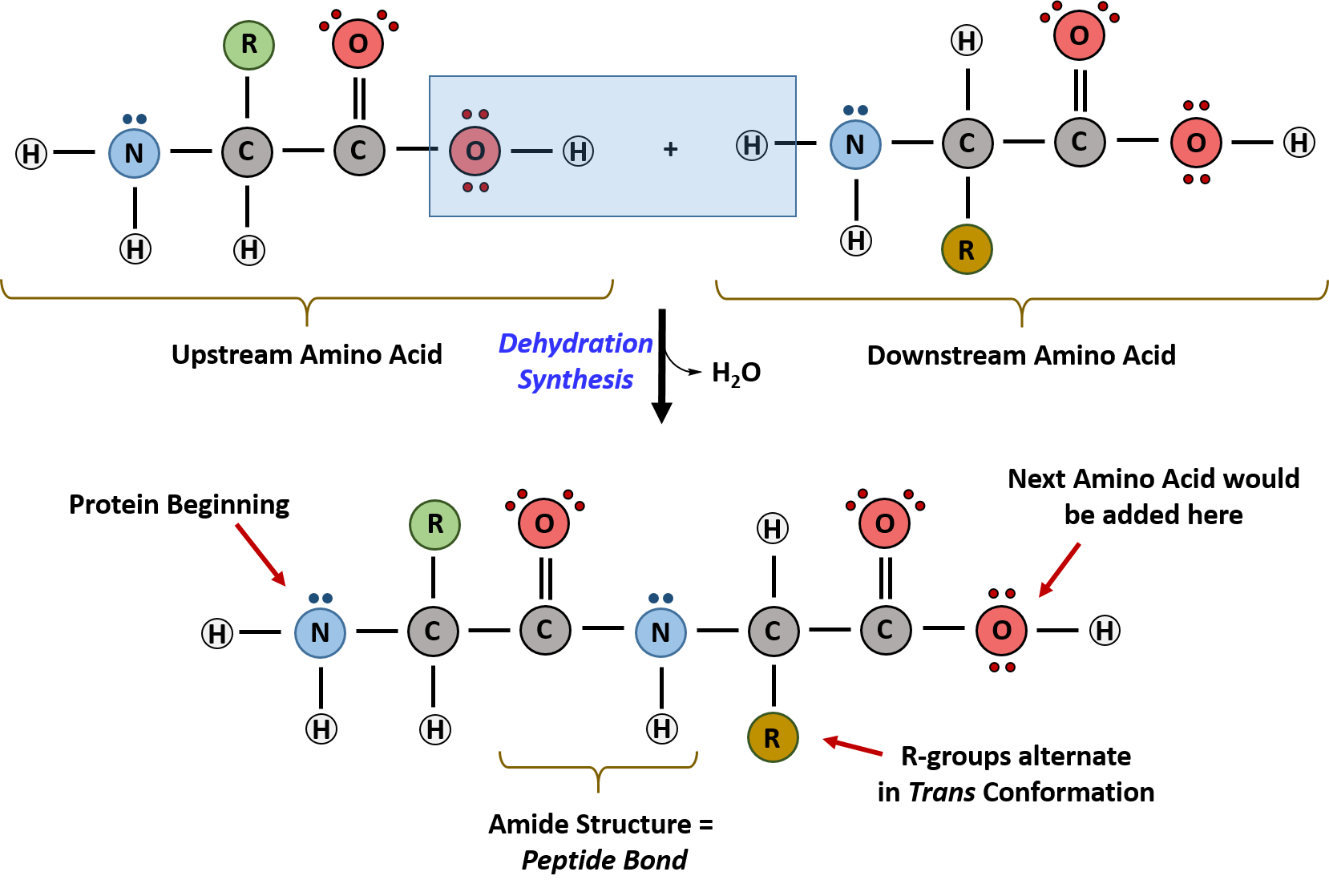
Results of combinations offrameshift mutations show that the code is in triplets. 64 different combinations offour nucleotides taken three at a time).Ģ. With three nucleotides, the set of allcombinations can encode If a codon were two nucleotides, theset of all combinations could encode onlyĬ. 20 amino acids are encoded bycombinations of 4 nucleotidesī. Three is the minimum number ofnucleotides per codon needed to encode 20 amino acids.Ī. tRNAs serve as anadaptor for translating from nucleic acid to proteinġ. The machinery for synthesizing proteins under the directionof template mRNA is the ribosome. it "speaks the language" of nucleic acids at one end and the"language" of proteins at the other end. A charged tRNA has an amino acid at oneend, and at the other end it has an anticodon for matching a codon in the mRNA ie. The adaptor molecule fortranslation is tRNA. Since there are 64 combinations of 4 nucleotides taken threeat a time and only 20 amino acids, the code is degenerate (more than one codon per amino acid, in mostcases). Eachgroup of three nucleotides encodes one amino acid. The nucleotidetriplet that encodes an amino acid is called a codon. This demonstrated that the coding unit is 3nucleotides.

Experiments testing the effects offrameshift mutations showed that the deletion or addition of 1 or 2 nucleotidescaused a loss of function, whereas deletion or addition of 3 nucleotidesallowed retention of considerable function. The rules for translatingfrom the "language" of nucleic acids to that of proteins is the geneticcode. It must be translated into the encodedprotein. But the mature mRNA is not yet functional to the cell. Oncetranscription and processing of rRNAs, tRNAs and snRNAs are completed, the RNAsare ready to be used in the cell ‑ assembled into ribosomes or snRNPs andused in splicing and protein synthesis. Overview for Genetic Code and Translation:


 0 kommentar(er)
0 kommentar(er)
